













| General Information: |
|
| Name(s) found: |
gi|40795667
[NCBI NR]
gi|2656143 [NCBI NR] gi|119585377 [NCBI NR] gi|119585373 [NCBI NR] |
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Homo sapiens |
| Length: | 916 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAEGGGCRER PDAETQKSEL GPLMRTTLQR GAQWYLIDSR WFKQWKKYVG FDSWDMYNVG 60
61 EHNLFPGPID NSGLFSDPES QTLKEHLIDE LDYVLVPTEA WNKLLNWYGC VEGQQPIVRK 120
121 VVEHGLFVKH CKVEVYLLEL KLCENSDPTN VLSCHFSKAD TIATIEKEMR KLFNIPAERE 180
181 TRLWNKYMSN TYEQLSKLDN TVQDAGLYQG QVLVIEPQNE DGTWPRQTLQ SNGSGFSASY 240
241 NCQEPPSSHI QPGLCGLGNL GNTCFMNSAL QCLSNTAPLT DYFLKDEYEA EINRDNPLGM 300
301 KGEIAEAYAE LIKQMWSGRD AHVAPRMFKT QVGRFAPQFS GYQQQDSQEL LAFLLDGLHE 360
361 DLNRVKKKPY LELKDANGRP DAVVAKEAWE NHRLRNDSVI VDTFHGLFKS TLVCPECAKV 420
421 SVTFDPFCYL TLPLPLKKDR VMEVFLVPAD PHCRPTQYRV TVPLMGAVSD LCEALSRLSG 480
481 IAAENMVVAD VYNHRFHKIF QMDEGLNHIM PRDDIFVYEV CSTSVDGSEC VTLPVYFRER 540
541 KSRPSSTSSA SALYGQPLLL SVPKHKLTLE SLYQAVCDRI SRYVKQPLPD EFGSSPLEPG 600
601 ACNGSRNSCE GEDEEEMEHQ EEGKEQLSET EGSGEDEPGN DPSETTQKKI KGQPCPKRLF 660
661 TFSLVNSYGT ADINSLAADG KLLKLNSRST LAMDWDSETR RLYYDEQESE AYEKHVSMLQ 720
721 PQKKKKTTVA LRDCIELFTT METLGEHDPW YCPNCKKHQQ ATKKFDLWSL PKILVVHLKR 780
781 FSYNRYWRDK LDTVVEFPIR GLNMSEFVCN LSARPYVYDL IAVSNHYGAM GVGHYTAYAK 840
841 NKLNGKWYYF DDSNVSLASE DQIVTKAAYV LFYQRRDDEF YKTPSLSSSG SSDGGTRPSS 900
901 SQQGFGDDEA CSMDTN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
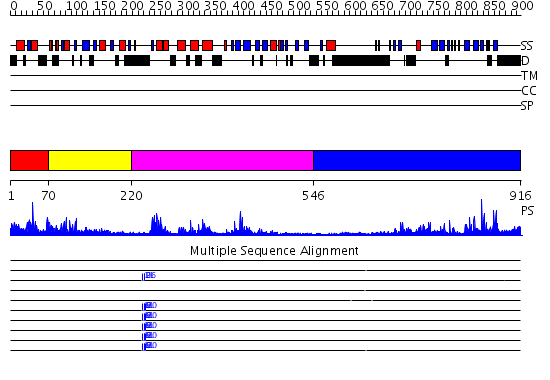
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..69] | 42.0 | Solution structure of the DUSP domain of hUSP15 | |
| 2 | View Details | [70..219] | 9.154902 | No description for 2g43A was found. | |
| 3 | View Details | [220..545] | 50.09691 | No description for 2gfoA was found. | |
| 4 | View Details | [546..916] | 51.522879 | No description for 2hd5A was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)