













| General Information: |
|
| Name(s) found: |
gi|9758035
[NCBI NR]
gi|21436231 [NCBI NR] gi|19423948 [NCBI NR] gi|15240689 [NCBI NR] |
| Description(s) found:
Found 12 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 806 amino acids |
Gene Ontology: |
|
| Cellular Component: |
vacuole
[IDA]
|
| Biological Process: | NONE FOUND |
| Molecular Function: |
alpha-N-acetylglucosaminidase activity
[ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MHSIKLVLLV LLIISFHSQT VSKHHPTIDG LLDRLDSLLP TSSVQESAAK GLLQRLLPTH 60
61 SQSFELRIIS KDACGGTSCF VIENYDGPGR IGPEILIKGT TGVEIASGLH WYLKYKCNAH 120
121 VSWDKTGGIQ VASVPQPGHL PRIDSKRIFI RRPVPWNYYQ NVVTSSYSYV WWGWERWERE 180
181 IDWMALQGIN LPLAFTGQEA IWQKVFKRFN ISKEDLDDYF GGPAFLAWAR MGNLHAWGGP 240
241 LSKNWLDDQL LLQKQILSRM LKFGMTPVLP SFSGNVPSAL RKIYPEANIT RLDNWNTVDG 300
301 DSRWCCTYLL NPSDPLFIEI GEAFIKQQTE EYGEITNIYN CDTFNENTPP TSEPEYISSL 360
361 GAAVYKAMSK GNKNAVWLMQ GWLFSSDSKF WKPPQLKALL HSVPFGKMIV LDLYAEVKPI 420
421 WNKSAQFYGT PYIWCMLHNF GGNIEMYGAL DSISSGPVDA RVSKNSTMVG VGMCMEGIEQ 480
481 NPVVYELTSE MAFRDEKVDV QKWLKSYARR RYMKENHQIE AAWEILYHTV YNCTDGIADH 540
541 NTDFIVKLPD WDPSSSVQDD LKQKDSYMIS TGPYETKRRV LFQDKTADLP KAHLWYSTKE 600
601 VIQALKLFLE AGDDLSRSLT YRYDMVDLTR QVLSKLANQV YTEAVTAFVK KDIGSLGQLS 660
661 EKFLELIKDM DVLLASDDNC LLGTWLESAK KLAKNGDERK QYEWNARTQV TMWYDSNDVN 720
721 QSKLHDYANK FWSGLLEDYY LPRARLYFNE MLKSLRDKKI FKVEKWRREW IMMSHKWQQS 780
781 SSEVYPVKAK GDALAISRHL LSKYFP |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
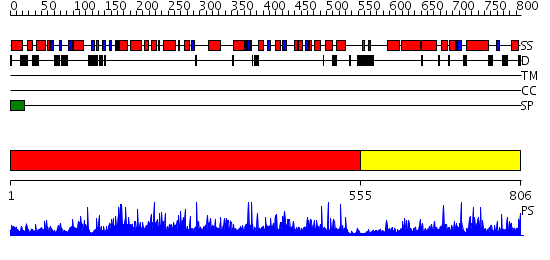
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..554] | 1.06 | beta-N-acetylhexosaminidase; beta-N-acetylhexosaminidase, N-terminal domain | |
| 2 | View Details | [555..806] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|